Dr. J. Nicholas Taylor, Specially Appointed Assistant Professor, Dr. Chun-Biu Li, Associate Professor, and Dr. Tamiki Komatsuzaki, Professor, along with their colleagues in the Laboratory of Molecule and Life Nonlinear Sciences, have successfully developed a new scheme for the extraction of microscopic states and energy landscapes from experimental single-molecule (SM) time-series. The new scheme uses methods from statistics and information theory to quantify errors in the time-series measurements and extract valuable and detailed information about biological systems.
The scheme relies on the appropriate treatment of errors that arise in SM measurements. To illustrate these errors, consider a series of fair coin tosses. If the coin were tossed an infinite number of times, exactly one half would land on heads and exactly one half on tails. Tossing the coin an infinite number of times is impossible of course, giving rise to finite sampling error. For example, if we have eight samples —that is, if the coin is tossed eight times— the number of heads may be five and the number of tails three. An additional eight tosses may yield five tails and three heads. Because the number of tosses is not infinite, we are uncertain as to what the exact percentage of the occurrence of heads and tails would be at the limit of infinite tosses, which is called finite sampling error. Now suppose that the coin is worn so that we cannot be completely certain as to whether a toss lands on heads or tails. This uncertainty in the measured value, that is, in the observation of heads or tails, is measurement error. The consequence of these errors is that we can never predict, with complete certainty, exactly how many heads or tails we will observe in a given set of tosses. Fortunately though, we can quantify these errors through the use of statistical methods, thus we can determine how certain we are about our predicted numbers of heads and tails.
In SM time-series experiments, an observable quantity, such as a number of photons or an intra-molecular distance, is measured in discrete increments of time. Each measurement, as in the case of a coin that is tossed a finite number of times and is worn so that heads and tails cannot be distinguished with certainty, contains measurement error and finite sampling error. After the errors are quantified, extracting the properties of the system underlying the measurements becomes the objective. To this end, methods of information theory are used to classify each measurement into a microscopic state. Microscopic states are analogous to the sides of the coin, except that the coin can have more than just two sides and the probabilities of landing on heads or tails can be different from one half. Various properties are computed, including each state’s occupation probabilities, which are analogous to the numbers of heads or tails in a set of tosses, the free energy profile, which is a depiction of the forces at work that would cause a tossed coin to land on either heads or tails, and state-to-state connectivity, which is analogous to the probabilities of the next toss landing heads or tails after the previous toss landed heads. These properties contain valuable information about the processes and forces that drive the system’s observed behavior.
The new method was applied to experimental SM time-series acquired for a neurological proteinknown as the AMPA receptor, which mediate the transmission of Calcium ion through the central nervous system. As consequence, it was uncovered how states are organized in the AMPA receptor and how it closely relates to the amount of the transmitted Calcium ion. They are the most abundant proteins in our nervous systems and have been implicated in disorders such as depression and epilepsy. Uncovering vital information about the AMPA receptor has deepened understanding of how the protein moves in response to various biological stimuli, which is also expected to be applicable for medical applications.
This research was conducted in collaboration with the research group of Prof. Christy F. Landes (Department of Chemistry, Rice University, Houston, TX, U.S.A.) and was published in Scientific Reports on 17th March 2015, volume 5, pages 9174-9182.
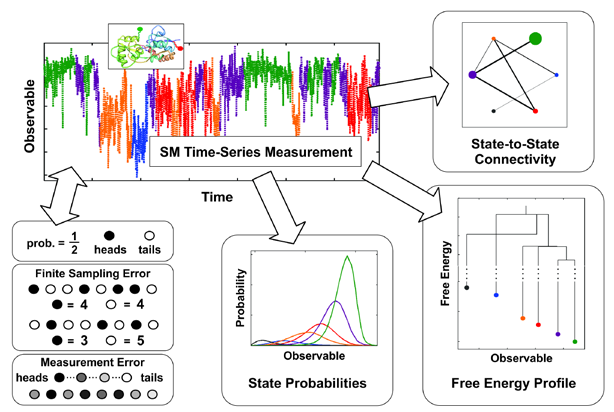
Figure. A SM time-series measurement is shown in the upper left along with a depiction of the AMPA receptor. Illustrations of finite sampling and measurement errors are shown to the bottom left. Quantification of these errors allows the measurements to be classified into microscopic states, which subsequently allows for the calculation of state probabilities (bottom center), free energy profiles (bottom right), and state-to-state connectivity (top left), leading to a detailed depiction of the processes and forces driving the system under observation.










